NAKAI Lab.
Laboratory of Functional Analysis in silico
Human Genome Center
The Institute of Medical Science
The University of Tokyo
resource

|
PSORT: Prediction of protein localization sites in cells A computer program for the prediction of protein localization sites in cells. It receives the information of an amino acid sequence and its source orgin, e.g., Gram-negative bacteria, as inputs. Then, it analyzes the input sequence by applying the stored rules for various sequence features of known protein sorting signals. Finally, it reports the possiblity for the input protein to be localized at each candidate site with additional information. |
|
Melina2: A web-based promoter analysis tool Melina2 is a tool which finds potential DNA motifs with a user friendly interface. It runs a maximum of 4 programs from 5 motif prediction tools (Consensus, MEME, Gibbs Sampler, MDScan, Weeder), and returns graphical representations of their results. |
|
DBTSS: Database of Transcriptional Start Sites DBTSS represents exact positions of transcriptional start sites (TSSs) in the genome based on our unique experimentally validated TSS sequencing method (TSS-seq). |
|
DBTGR: Database of Tunicate Gene Regulation Tunicates are lower chordates and share basic gene repertoires and many characteristics with vertebrates. DBTGR provides information on tunicate gene regulation such as the location of expression, or the identified regulatory elements present in promoter sequences. |

|
DBTBS: A database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information DBTBS is a reference database on transcriptional regulation in Bacillus subtilis, summarizing the experimentally characterized transcription factors, their recognition sequences and the genes they regulate. |
|
HINTdb: Homologous Interactions Database HINTdb is a protein-protein interaction database which contains interactions from 105 species with their homologs in one or more species. The protein-protein interaction data includes that from small scale as well as high-throughput experiments from IntAct, BioGRID, HPRD, DIP and MINT. The homologous interactions are also derived from these databases. |
|
HitPredict: A database of high confidence protein-protein interactions HitPredict is a resource of experimentally determined protein-protein interactions with reliability scores. Protein-protein interactions from IntAct, BioGRID, HPRD, MINT and DIP are combined, annotated and scored. The reliability score is calculated based on the experimental details of each interaction and the sequence, structure and functional annotations of the interacting proteins. |

|
TimeXNet: identifying active gene sub-networks using time-course gene expression profiles TimeXNet is an application that predicts activated pathways during a cellular response using time-course gene expression profiles in the context of a large molecular interaction network. TimeXNet Web: Identifying cellular response networks from diverse omics time-course data TimeXNet Web supports the analysis of high-throughput data from multiple species by providing high quality, weighted protein-protein interaction networks. |
|
DBTMEE: Database of transcriptome in mouse eary embryos DBTMEE is a free database designed to manipulate gene expression information from our ultralarge-scale whole-transcriptome analysis of mouse early embryos. Since integrative approaches with multiple public analytical data have become indispensable for studying embryogenesis, we combined the gene expression profile with various public resources. |
|
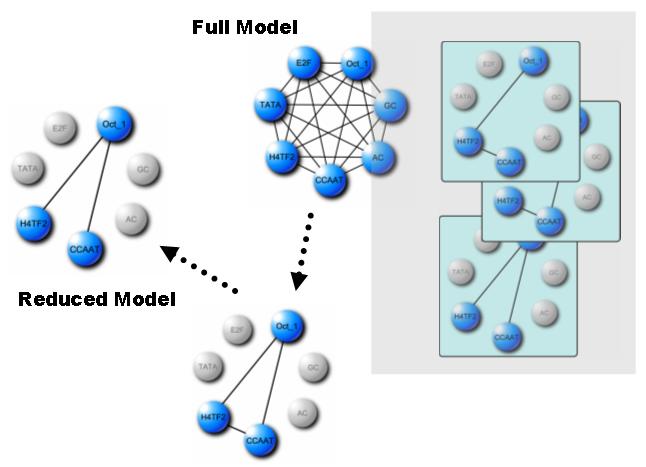
LLM (Log-Linear Model): inferring probabilistic conditional independency from combinatorial regulation of transcription factors LLM deals with spurious interactions of given variables (e.g. transcription factor bindings) that may lead deceptive inferences. Given discrete (0 or 1) input data, this method uses a log-linear modeling that deals with probabilistic conditional independency for detecting spuriousness. |
|
OpenTein: Open Teratoma Investigation OpenTein is a searchable, zoomable and annotatable web-based repository system that enables to archive and freely distribute high-resolution whole-slide images and relevant records for the stem cell-derived teratoma formation assay. |
|
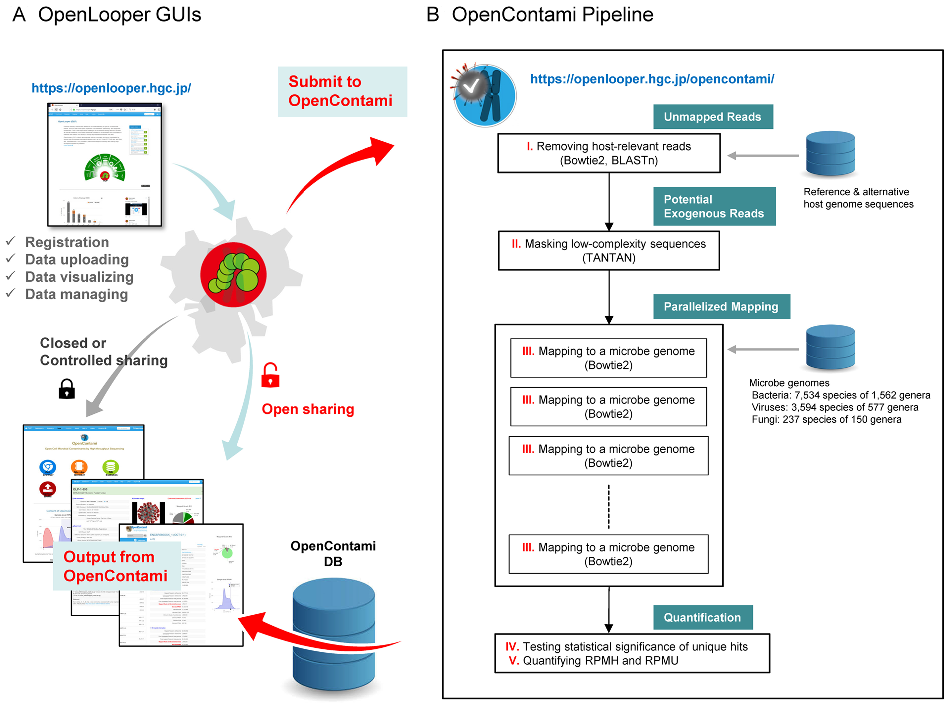
OpenContami: A web-based application for detecting microbial contaminants in next-generation sequencing data OpenContami (OCT) investigates the origin of sequenced reads from user-uploaded BAM files and provides highly probable microbial contaminants present in the BAM files that may be contaminated by the laboratory reagents, sequencer carryovers, cross-contamination, etc. |
|
RegMed: Open source tools for regenerative medicine RegMed is developing public databases and open source software for open science, which includes OpenIAV (Open Image Annotation Viewer), OpenSOP (Open Standard Operating Procedure). |